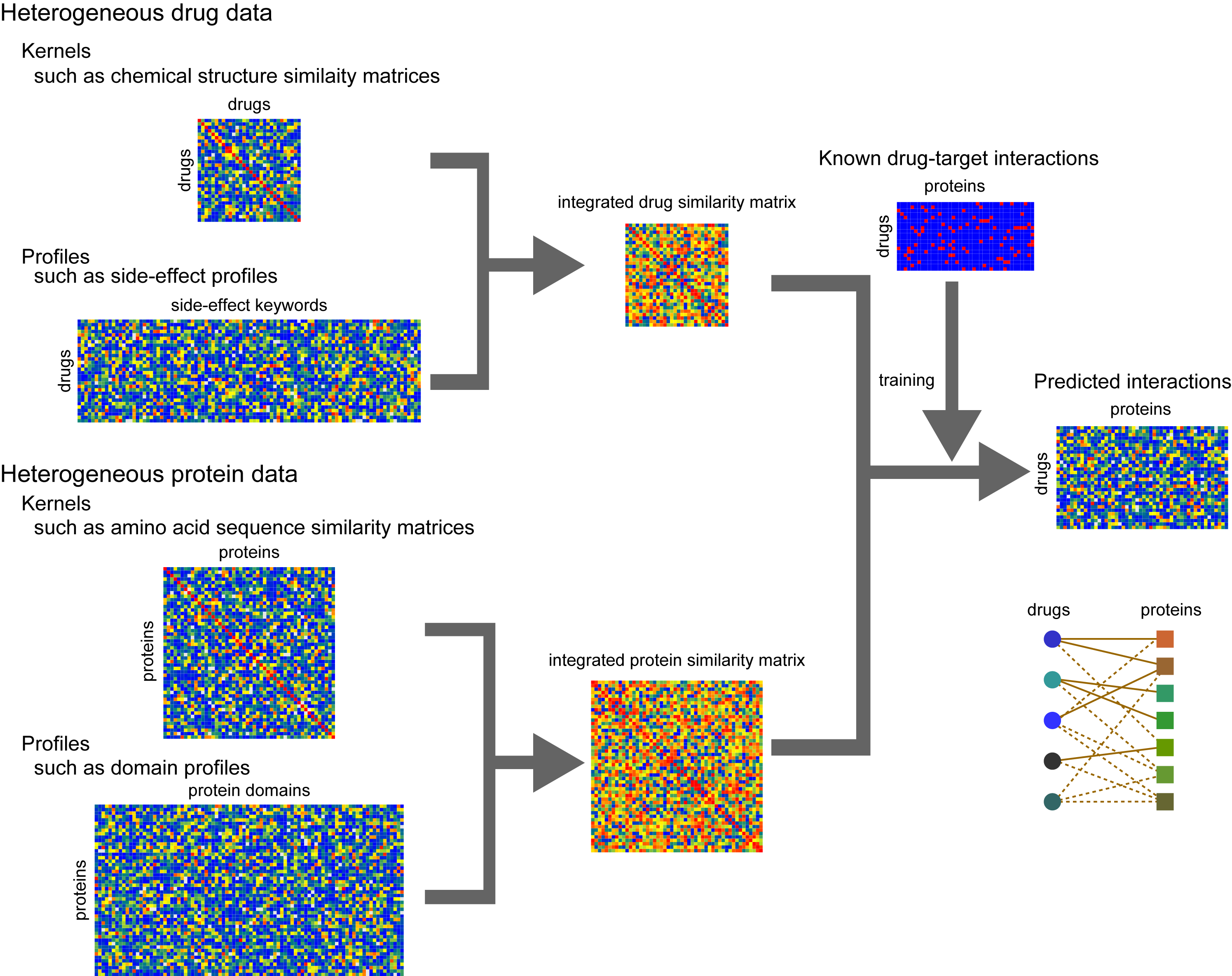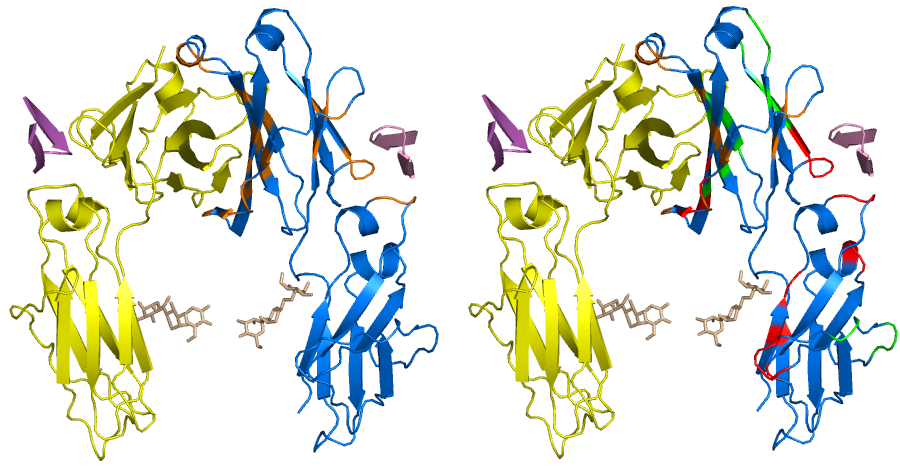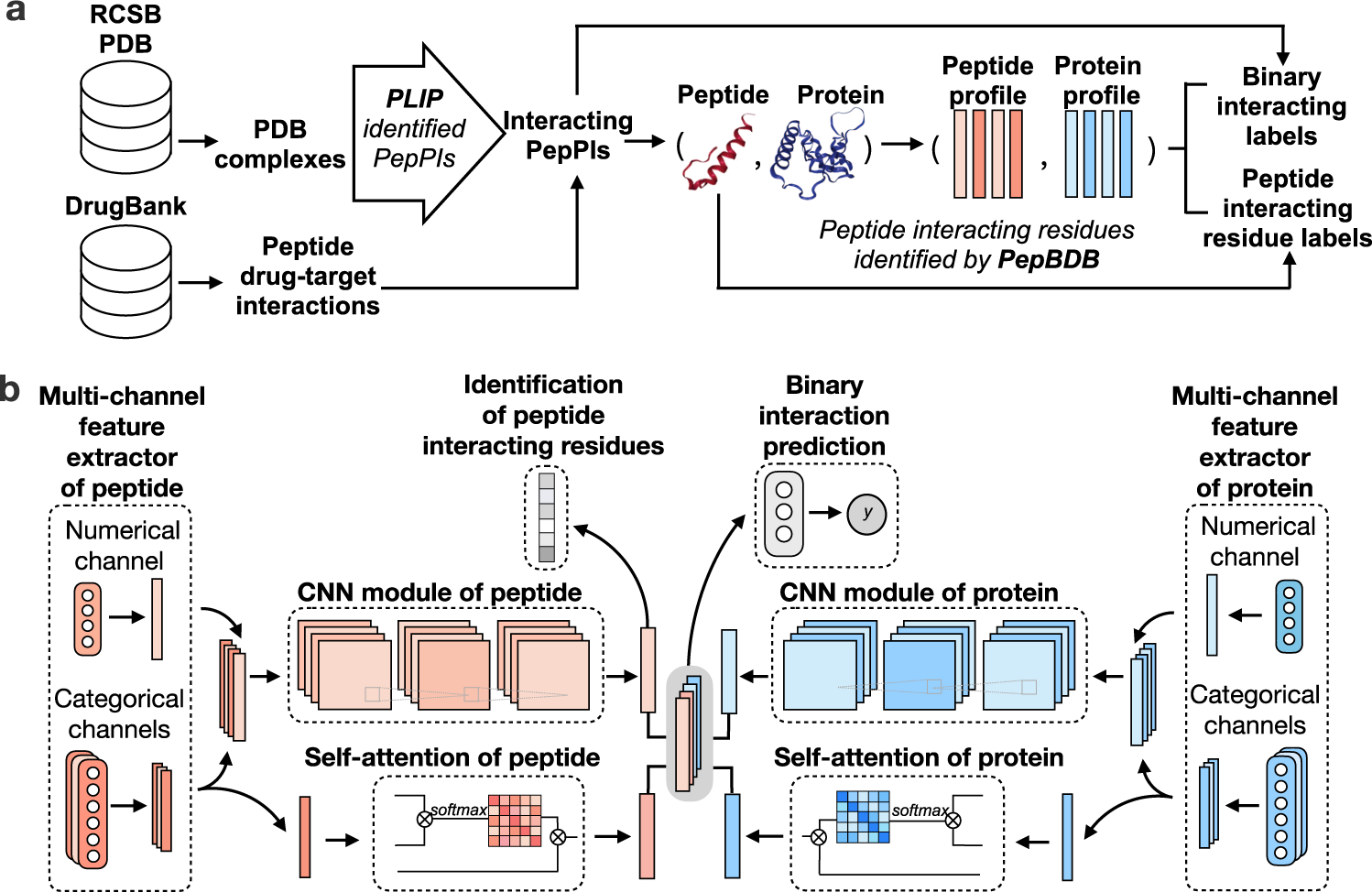
A deep-learning framework for multi-level peptide–protein interaction prediction | Nature Communications
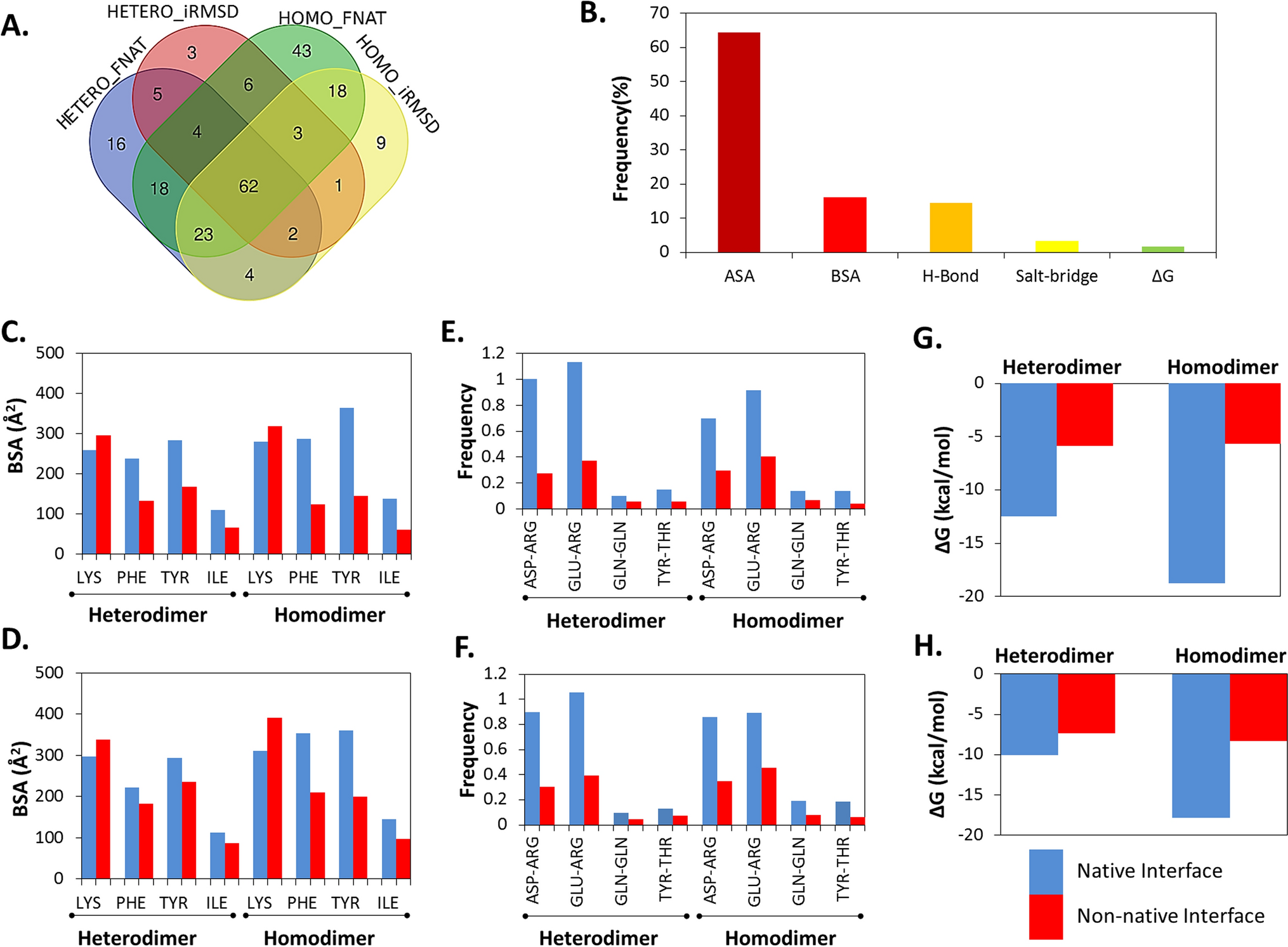
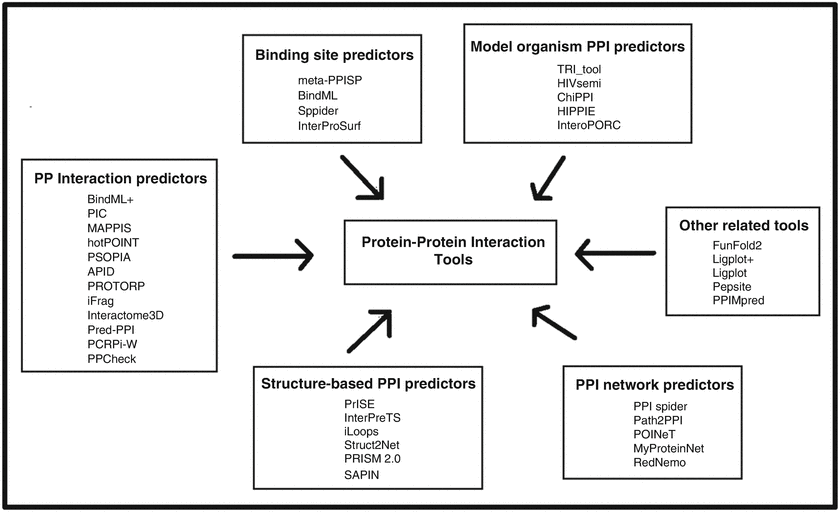
Classification and prediction of protein–protein interaction interface using machine learning algorithm | Scientific Reports
Plantae | Proteome-wide, structure-based prediction of protein-protein interactions (Plant Physiol) | Plantae
Frontiers | LZerD Protein-Protein Docking Webserver Enhanced With de novo Structure Prediction | Molecular Biosciences
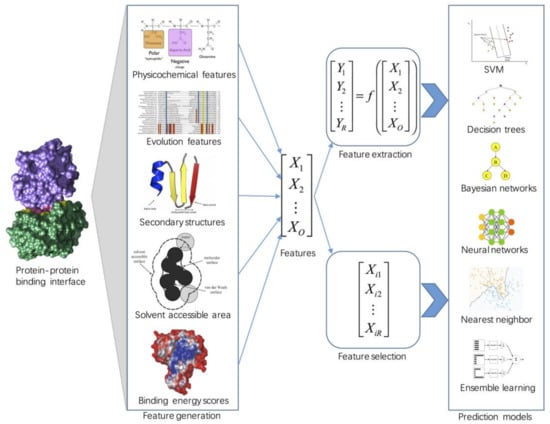
Molecules | Free Full-Text | Machine Learning Approaches for Protein–Protein Interaction Hot Spot Prediction: Progress and Comparative Assessment | HTML
Frontiers | In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions | Molecular Biosciences
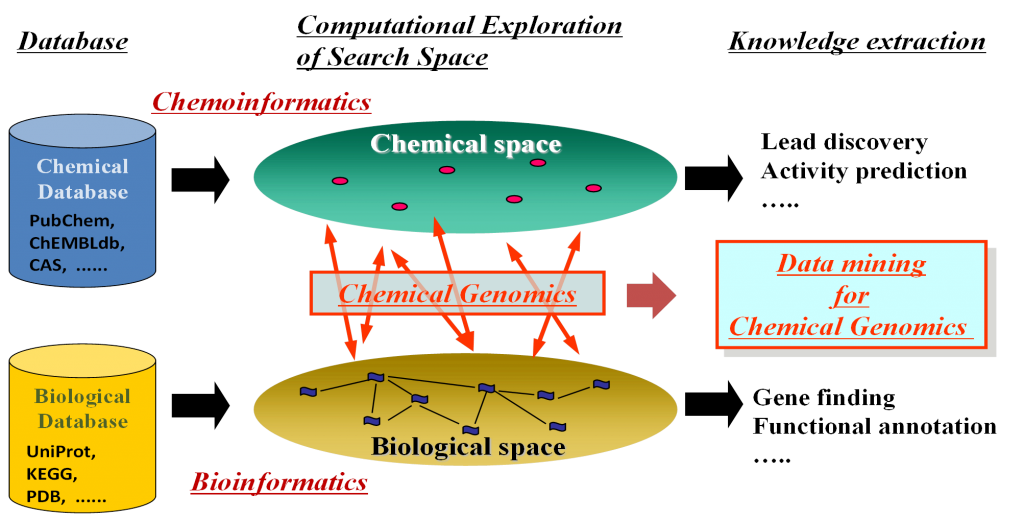
In silico target prediction for protein-protein interaction (PPI) small molecule inhibitors using CGBVS - In Silico 創薬

BindML: Software for Predicting Protein-Protein Interaction Sites using Phylogenetic Substitution Models
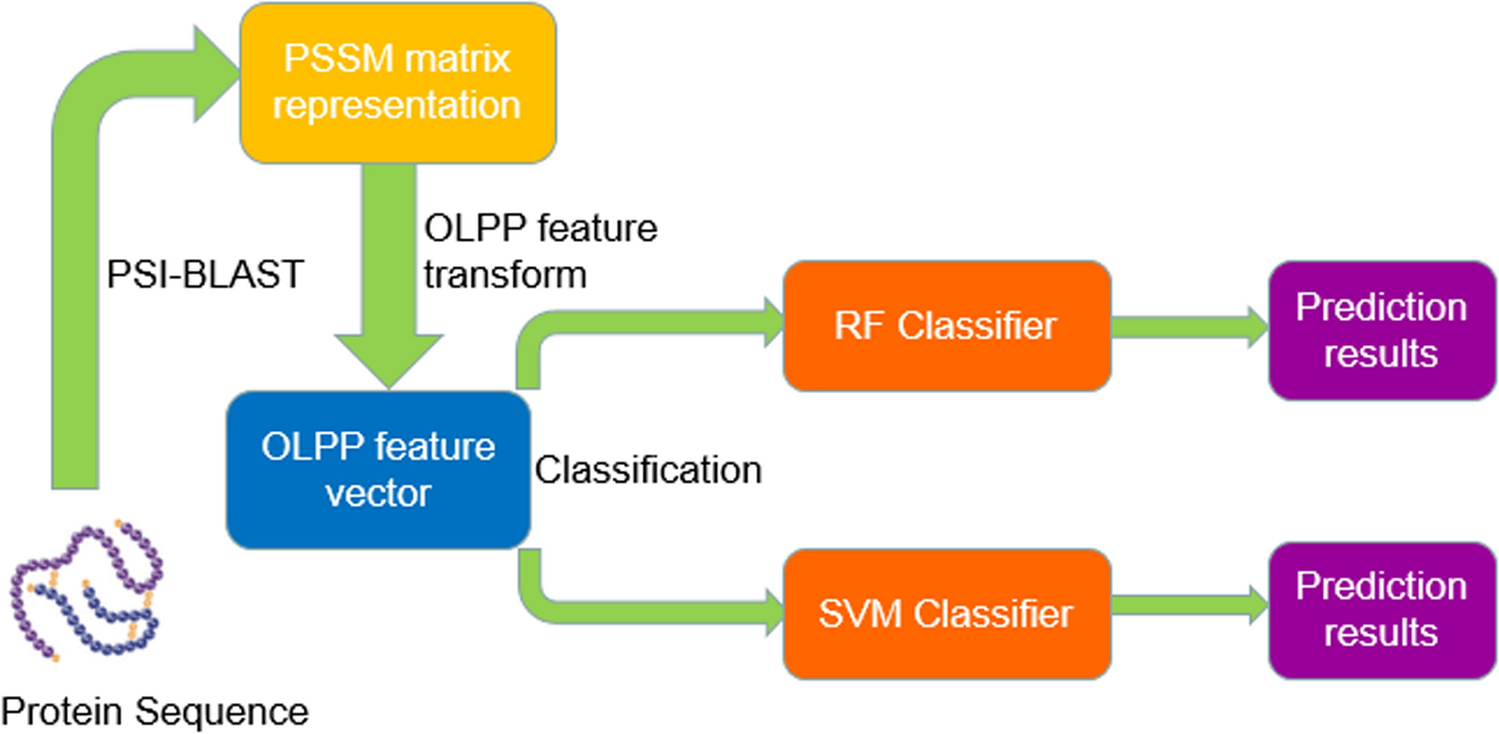
Robust and accurate prediction of protein–protein interactions by exploiting evolutionary information | Scientific Reports
SFPEL-LPI: Sequence-based feature projection ensemble learning for predicting LncRNA-protein interactions
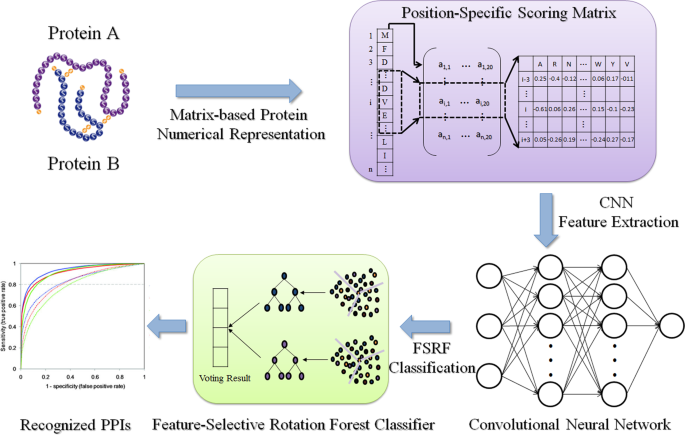
Predicting Protein-Protein Interactions from Matrix-Based Protein Sequence Using Convolution Neural Network and Feature-Selective Rotation Forest | Scientific Reports
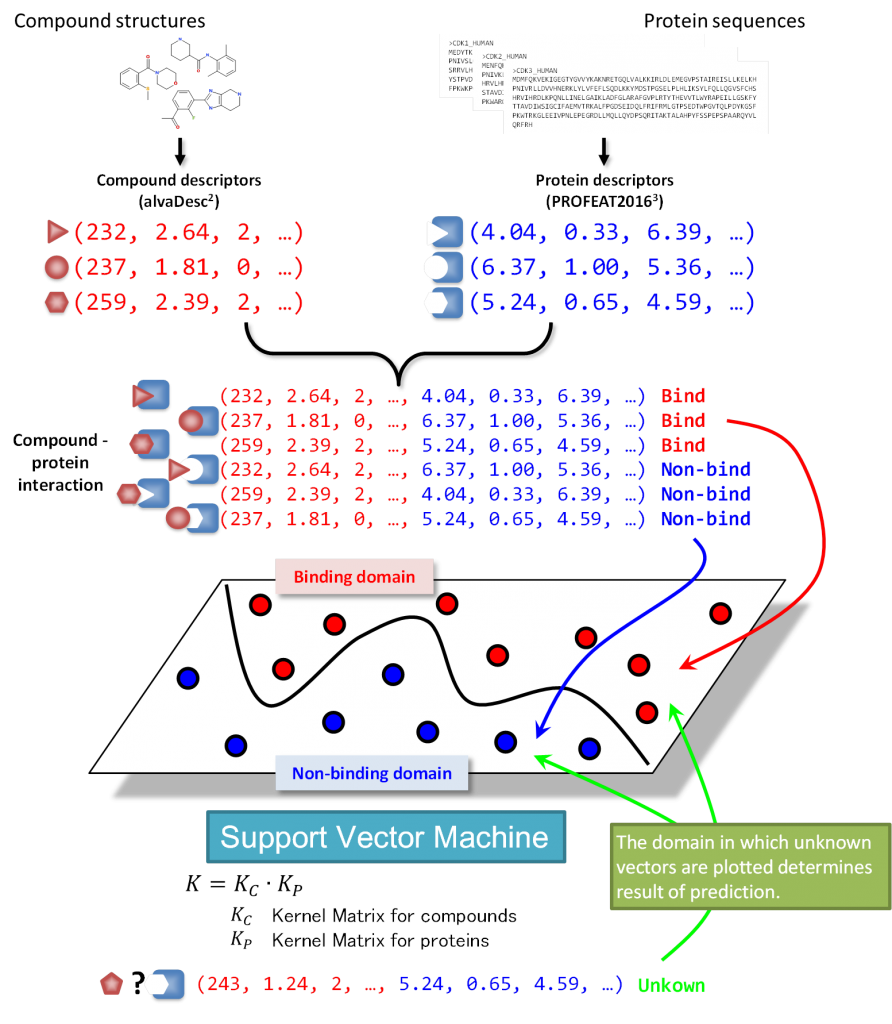
In silico target prediction for protein-protein interaction (PPI) small molecule inhibitors using CGBVS - In Silico 創薬

BindML: Software for Predicting Protein-Protein Interaction Sites using Phylogenetic Substitution Models
SPServer: split-statistical potentials for the analysis of protein structures and protein–protein interactions | BMC Bioinformatics | Full Text

Protein-assisted RNA fragment docking (RnaX) for modeling RNA–protein interactions using ModelX | PNAS
PLOS ONE: Protein-Protein Interaction Site Predictions with Three-Dimensional Probability Distributions of Interacting Atoms on Protein Surfaces